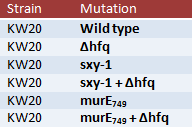
To do this, I'll be growing stock of the following cells:
To clarify, Δhfq is the hfq knockout. sxy-1 is a point mutation in the sxy (tfoX) gene that causes hypercompetence, thought to be due to changes in the secondary structure of sxy mRNA that normally inhibits translation of the mRNA (and thus normally inhibits competence). murE749 is different point mutation in the murE gene (responsible for a step in peptidoglycan synthesis) and oddly enough also causes hypercompetence when mutated. The data from the murE mutations will likely not be used in this experiment, but it will perhaps become more useful later as we learn more about murE and hfq.
I will be taking all these cell lines and subjecting them to the following conditions and measuring transformation frequency:
Here is a breakdown of the experimental logic.
General question: Why does knocking out hfq decrease a culture's transformation frequency?
Theories:
- The lack of hfq somehow stabilizes sxy mRNA and lowers transformation frequency.
- The lack of hfq prevents spikes of cAMP concentration that are required to induce competence.
- Both 1. and 2.
- Neither 1. or 2.
Predictions under each theory: (underline = prediction)
- if hfq is needed to destabilize sxy mRNA as theory 1. would have it, then the sxy-1 mutation would allow destabilization of the RNA strand independent of hfq thus would allow "recovery" of the lack of hfq and have high transformation frequency in all cases
- if Δhfq reduces cAMP production (theory 2.) then adding cAMP should make up for the transformation frequency defect but we would not expect recovery when extra cAMP is not given
- theory 3. would say that the phenotype is going to be whatever the most limiting phenotype is predicted from theory 1. and 2.
- theory 4. cannot make any obvious predictions